Research
Our group develops innovative methods for studying the structure and dynamics of biomacromolecules. These include bioinformatics approaches to exploit the relationship between a sequence and a structure as well as coarse grained and multiscale modelling. The current research is focused on:
Computational analyses of biomolecules
BioShell software package, that has been started in 2006, is a comprehensible set of tools for structural bioinformatics. The most recently published version 3.0 of the package provides Python bindings to C++ modules of BioShell.
Interatomic interactions
The newest version allows also to detect and analyse interatomic interactions, such as hydrogen bonds, Van der Waals and stacking interactions.
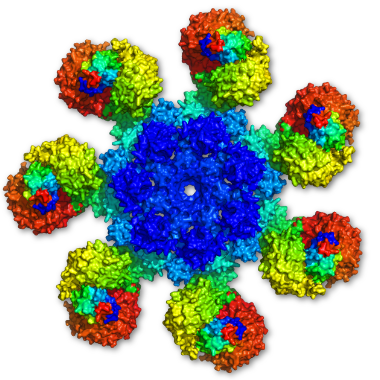
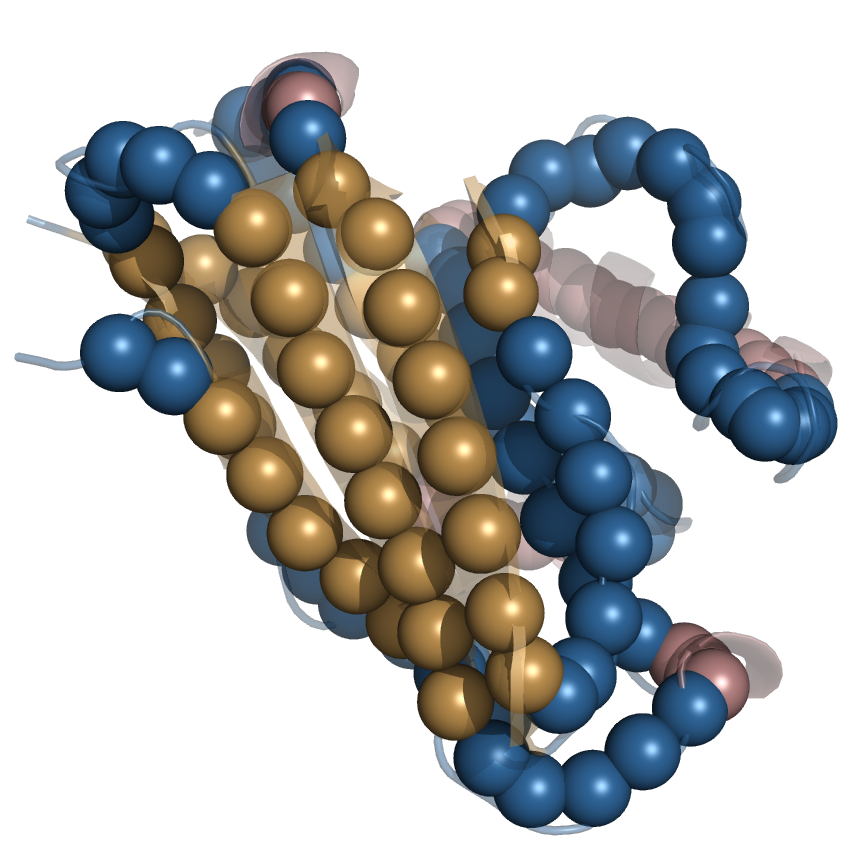
Coarse grained modelling of proteins
In coarse-grained models, a group of atoms , such as an amino acid side chain, a residue or a small molecule, is substituted with a single interaction center – united atom. Numerous models have been proposed in the past few decades, that differ in how they represent all-atom molecules and how they describe interactions.
SURPASS model
in the Single United Residue per Pre-Averaged Secondary Structure fragment (SURPASS) model a single coarse-grained atom corresponds roughly to an amino acid residue.
Project has been supported by:
2018/29/B/ST6/01989 NCN research grant:
,,Novel combination of Rosetta method with SURPASS coarse grained model into a multiscale protocol for modelling proteins and their complexes''.
![]()
The project has been realized at Warsaw University.




